Extending ergmito
George G. Vega Yon
2020-08-09
Source:vignettes/extending-ergmito.Rmd
extending-ergmito.RmdThe ergmito’s workhorse are two other functions: (1) ergm’s ergm.allstats which is used to compute the support of model’s sufficient statistics, and (2) ergmito_formulae which is a wrapper of that same function, and that returns a list including the following two functions: loglike and grad, the functions to calculate the joint log-likelihood of the model and its gradient.
library(ergmito) data(fivenets) model_object <- ergmito_formulae(fivenets ~ edges + ttriad) # Printing the model object model_object #> ergmito log-likelihood function #> Number of networks: 5 #> Model: fivenets ~ edges + ttriad #> Available elements by using the $ operator: #> loglik: function (params, ..., as_prob = FALSE, total = TRUE) grad : function (params, ...) # Printing the log-likelihood function model_object$loglik #> function (params, ..., as_prob = FALSE, total = TRUE) #> { #> if (total) { #> ans <- sum(exact_loglik(ergmito_ptr, params = params, #> ..., as_prob = as_prob)) #> if (!as_prob && !is.finite(ans)) #> return(-.Machine$double.xmax * 1e-100) #> else return(ans) #> } #> else { #> exact_loglik(ergmito_ptr, params = params, ..., as_prob = as_prob) #> } #> } #> <bytecode: 0x72fe538> #> <environment: 0x72f8d60>
Besides of the log-likelihood function and the gradient function, ergmito_formulae also returns We can take a look at each component from our previous object:
# The vectors of weights str(model_object$stats_weights) #> List of 5 #> $ : num [1:40] 66 12 168 72 48 196 120 1 88 1 ... #> $ : num [1:40] 66 12 168 72 48 196 120 1 88 1 ... #> $ : num [1:40] 66 12 168 72 48 196 120 1 88 1 ... #> $ : num [1:40] 66 12 168 72 48 196 120 1 88 1 ... #> $ : num [1:40] 66 12 168 72 48 196 120 1 88 1 ... # The matrices of the sufficient statistics str(model_object$stats_statmat) #> List of 5 #> $ : num [1:40, 1:2] 2 1 4 8 7 3 6 12 6 0 ... #> ..- attr(*, "dimnames")=List of 2 #> .. ..$ : NULL #> .. ..$ : chr [1:2] "edges" "ttriple" #> $ : num [1:40, 1:2] 2 1 4 8 7 3 6 12 6 0 ... #> ..- attr(*, "dimnames")=List of 2 #> .. ..$ : NULL #> .. ..$ : chr [1:2] "edges" "ttriple" #> $ : num [1:40, 1:2] 2 1 4 8 7 3 6 12 6 0 ... #> ..- attr(*, "dimnames")=List of 2 #> .. ..$ : NULL #> .. ..$ : chr [1:2] "edges" "ttriple" #> $ : num [1:40, 1:2] 2 1 4 8 7 3 6 12 6 0 ... #> ..- attr(*, "dimnames")=List of 2 #> .. ..$ : NULL #> .. ..$ : chr [1:2] "edges" "ttriple" #> $ : num [1:40, 1:2] 2 1 4 8 7 3 6 12 6 0 ... #> ..- attr(*, "dimnames")=List of 2 #> .. ..$ : NULL #> .. ..$ : chr [1:2] "edges" "ttriple" # The target statistic model_object$target_stats #> edges ttriple #> [1,] 2 0 #> [2,] 7 4 #> [3,] 4 0 #> [4,] 5 2 #> [5,] 2 0
All this is closely related to the output object from the function ergm.allstats. The next section shows how all this works together to estimate the model parameters using Metropolis-Hastings MCMC.
Example: Bayesian inference with fivenets
Suppose that we have a prior regarding the distribution of the fivenets model: The edges parameter is normally distributed with mean -1 and variance 2, while the nodematch("female") term has the same distribution but with mean 1. We can implement this model using a Metropolis-Hastings ratio. First, we need to define the log of the posterior distribution:
# Analyzing the model model_object <- ergmito_formulae(fivenets ~ edges + nodematch("female")) #> Warning: `set_attrs()` is deprecated as of rlang 0.3.0 #> This warning is displayed once per session. # Defining the logposterior logposterior <- function(p) { model_object$loglik(params = p) + sum(dnorm(p, mean = c(-1,1), sd = sqrt(2), log = TRUE)) }
For this example, we are using the fmcmc R package. Here is how we put everything together:
# Loading the required R packages library(fmcmc) library(coda) # Is it working? logposterior(c(-1, 1)) #> [1] -38.24283 # Now, calling the MCMC function from the fmcmc R package ans <- MCMC( fun = logposterior, initial = c(0, 0), # 5,000 steps sampling one of every ten iterations nsteps = 5000, thin = 10, # We are using a normal transition kernel with .5 scale and updates are done # one variable at a time in a random order kernel = kernel_normal(scale = .5, scheme = "random") )
We can now visualize our results. Since the resulting object is of class mcmc.list, which is implemented in the coda R package for MCMC diagnostics, we can use all the methods included in the package:
plot(ans)
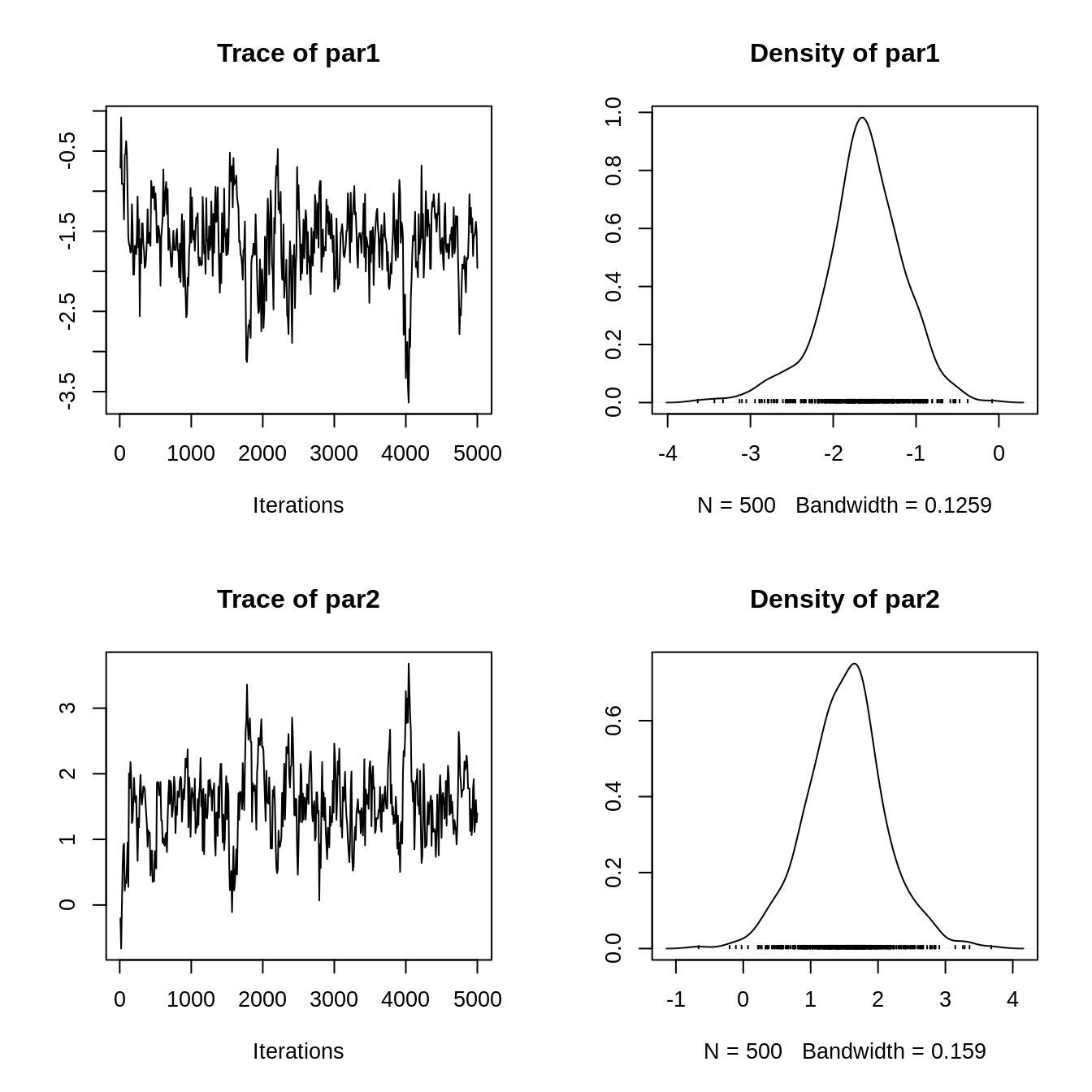
summary(ans) #> #> Iterations = 10:5000 #> Thinning interval = 10 #> Number of chains = 1 #> Sample size per chain = 500 #> #> 1. Empirical mean and standard deviation for each variable, #> plus standard error of the mean: #> #> Mean SD Naive SE Time-series SE #> par1 -1.636 0.4872 0.02179 0.05699 #> par2 1.517 0.5813 0.02600 0.06940 #> #> 2. Quantiles for each variable: #> #> 2.5% 25% 50% 75% 97.5% #> par1 -2.7822 -1.881 -1.624 -1.329 -0.7182 #> par2 0.3567 1.158 1.540 1.855 2.7509
Finally, we can compare this result with what we obtain from the ergmito function
summary(ergmito(fivenets ~ edges + nodematch("female"))) #> Warning: The observed statistics (target.statistics) are near or at the boundary #> of its support, i.e. the Maximum Likelihood Estimates maynot exist or be hard to #> be estimated. In particular, the statistic(s) "edges", "nodematch.female". #> #> ERGMito estimates (MLE) #> This model includes 5 networks. #> #> formula: #> fivenets ~ edges + nodematch("female") #> #> Estimate Std. Error z value Pr(>|z|) #> edges -1.70475 0.54356 -3.1363 0.001711 ** #> nodematch.female 1.58697 0.64305 2.4679 0.013592 * #> --- #> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 #> AIC: 73.34109 BIC: 77.52978 (Smaller is better.)