Goodness of Fit diagnostics for ERGMito models
gof_ergmito( object, GOF = NULL, GOF_update = NULL, probs = c(0.05, 0.95), sim_ci = FALSE, R = 50000L, ncores = 1L, ... ) # S3 method for ergmito_gof plot( x, y = NULL, main = NULL, sub = NULL, tnames = NULL, sort_by_ci = FALSE, ... )
Arguments
| object | An object of class ergmito. |
|---|---|
| GOF | Formula. Additional set of parameters to perform the GOF. |
| GOF_update | Formula. See the section on model updating in |
| probs | Numeric vector. Quantiles to plot (see details). |
| sim_ci | Logical scalar. If |
| R | Integer scalar. Number of simulations to generate (passed to sample).
This is only used if |
| ncores | Integer scalar. Number of cores to use for parallel computations (currently ignored). |
| ... | Further arguments passed to stats::quantile. |
| x | An object of class |
| y | Ignored. |
| main, sub | Title and subtitle of the plot (see graphics::title). |
| tnames | A named character vector. Alternative names for the terms. |
| sort_by_ci | Logical scalar. When |
Value
An object of class ergmito_gof. This is a list with the following
components:
ciA list of matrices of lengthnnets(object)with the corresponding confidence intervals for the statistics of the model.target_statsA matrix of the target statistics.ergmito.probsA list of numeric vectors of lengthnnets(object)with the probabilities associated to each possible structure of network.probsThe value passed viaprobs.modelThe fitted model.term_namesCharacter vector. Names of the terms used in the model.quantile.argsA list of the values passed via....
Details
The Goodness of Fit function uses the fitted ERGMito to calculate a given confidence interval for a set of sufficient statistics. By default (and currently the only available option), this is done on the sufficient statistics specified in the model.
In detail, the algorithm is executed as follow:
For every network in the list of networks do:
Calculate the probability of observing each possible graph in its support using the fitted model.
If
sim_ci = TRUE, drawRsamples from each set of parameters using the probabilities computed. Then using thequantilefunction, calculate the desired quantiles of the sufficient statistics. Otherwise, compute the quantiles using the analytic quantiles using the full distribution.'
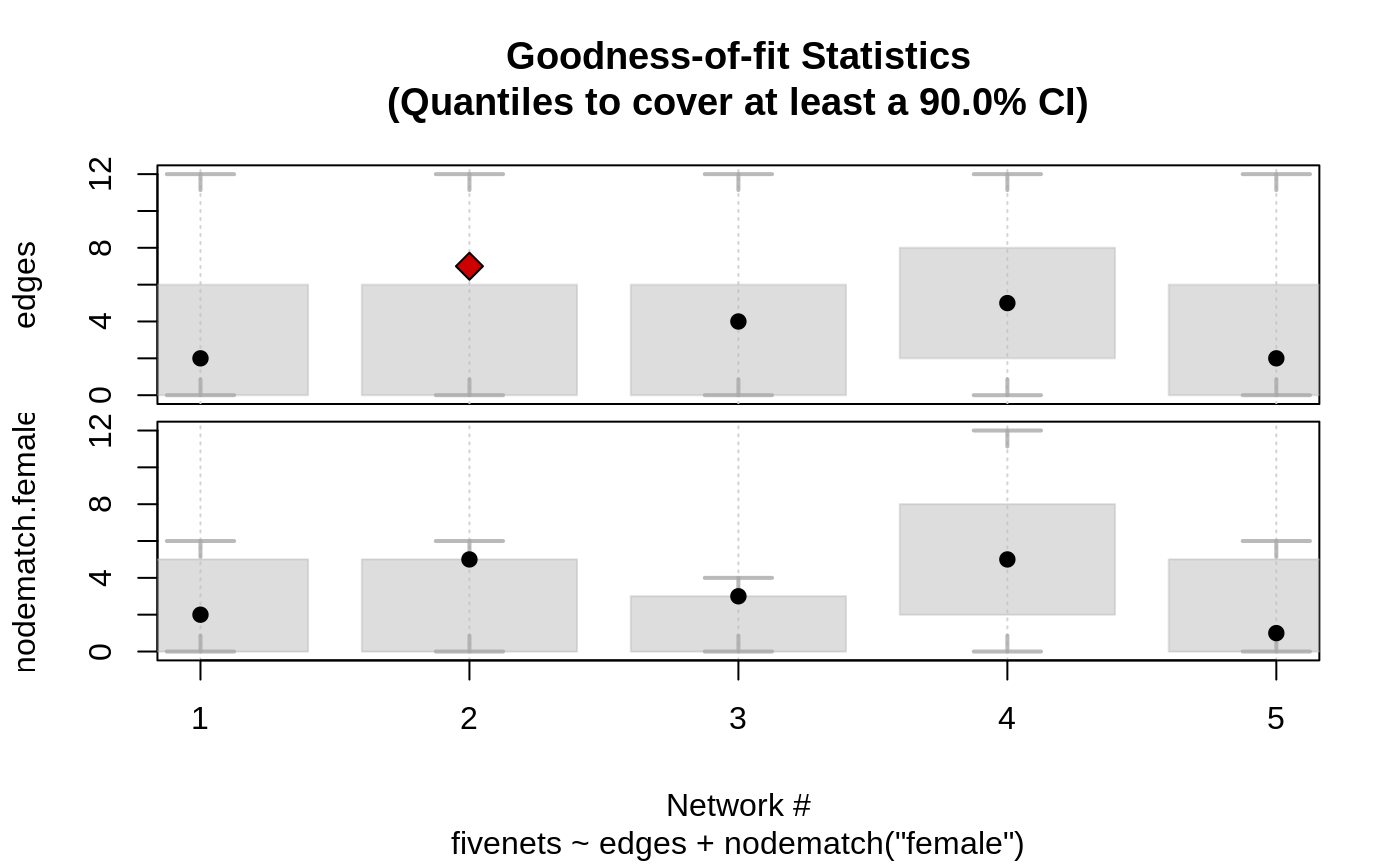
The plot method is particularly convenient since it graphically shows whether the target statistics of the model (observed statistics) fall within the simulated range.
The print method tries to copy (explicitly) the print method of the
gof function from the ergm R package.
Examples
# Fitting the fivenets model data(fivenets, package = "ergmito") fit <- ergmito(fivenets ~ edges + nodematch("female"))#> Warning: The observed statistics (target.statistics) are near or at the boundary of its support, i.e. the Maximum Likelihood Estimates maynot exist or be hard to be estimated. In particular, the statistic(s) "edges", "nodematch.female".# Calculating the gof ans <- gof_ergmito(fit) # Looking at the results ans#> #> Goodness-of-fit for edges #> #> obs min mean max lower upper lower prob. upper prob. #> net 1 2 0 3.746607 12 0 6 0.008080856 0.9635261 #> net 2 7 0 3.746607 12 0 6 0.008080856 0.9635261 #> net 3 4 0 3.113122 12 0 6 0.020642814 0.9881036 #> net 4 5 0 5.647059 12 2 8 0.030934588 0.9514908 #> net 5 2 0 3.746607 12 0 6 0.008080856 0.9635261 #> #> #> Goodness-of-fit for nodematch.female #> #> obs min mean max lower upper lower prob. upper prob. #> net 1 2 0 2.823530 6 0 5 0.02201716 0.9891396 #> net 2 5 0 2.823530 6 0 5 0.02201716 0.9891396 #> net 3 3 0 1.882353 4 0 3 0.07855507 0.9509584 #> net 4 5 0 5.647059 12 2 8 0.03093459 0.9514908 #> net 5 1 0 2.823530 6 0 5 0.02201716 0.9891396 #> #> Note: Exact confidence intervals where used. This implies that the requestes CI may differ from the one used (see ?gof_ergmito). #>plot(ans)